Awesome
<p align="center"> <a href="https://github/openbiox/bioshiny"> <img alt="bioshiny" src="https://raw.githubusercontent.com/openbiox/bioshiny/master/doc/images/bioshiny-logo.svg?sanitize=true" width="200" /> </a> </p> <p align="center"> <a href="https://www.npmjs.com/package/bioshiny"><img src="https://img.shields.io/badge/lifecycle-experimental-orange.svg" alt="Life cycle: experimental"> <a href="https://circleci.com/gh/openbiox/bioshiny/tree/master"><img src="https://img.shields.io/circleci/project/github/openbiox/bioshiny/master.svg" alt="Build Status"></a> <a href="https://npmcharts.com/compare/bioshiny?minimal=true"><img src="https://img.shields.io/npm/dm/bioshiny.svg" alt="Downloads"></a> <a href="https://www.npmjs.com/package/bioshiny"><img src="https://img.shields.io/npm/v/bioshiny.svg" alt="Version"></a> <a href="https://www.npmjs.com/package/bioshiny"><img src="https://img.shields.io/npm/l/bioshiny.svg" alt="License"></a> </p>bioshiny is a project to develop the R Shiny applications for bioinformatics research.
bioshiny
- One-click to create bioshiny Shiny application and plugins
- Create and share more plugins.
- Using npm and GitHub to share the Shiny applications and plugins (CRAN team limited file size)
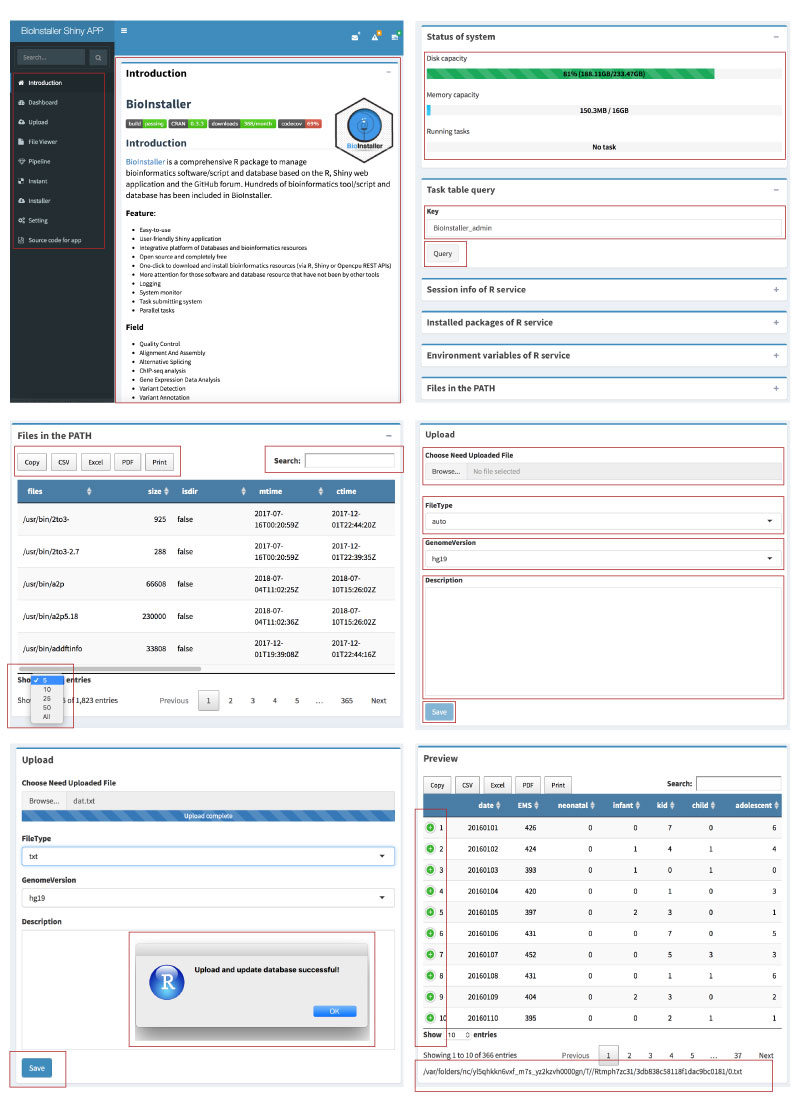
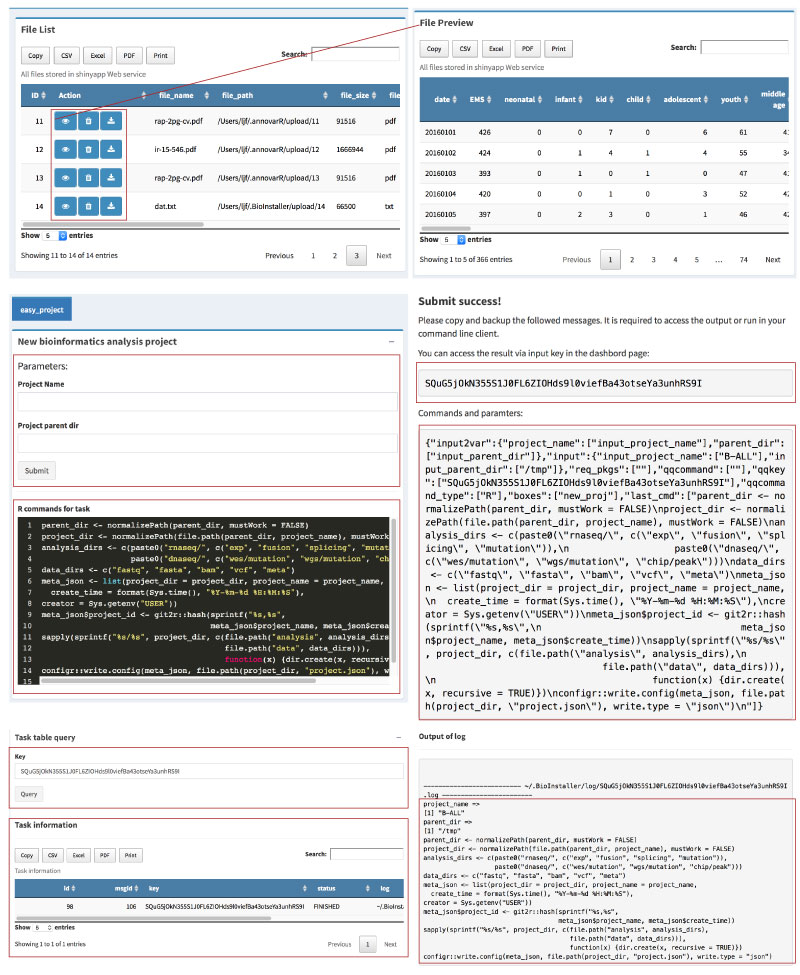
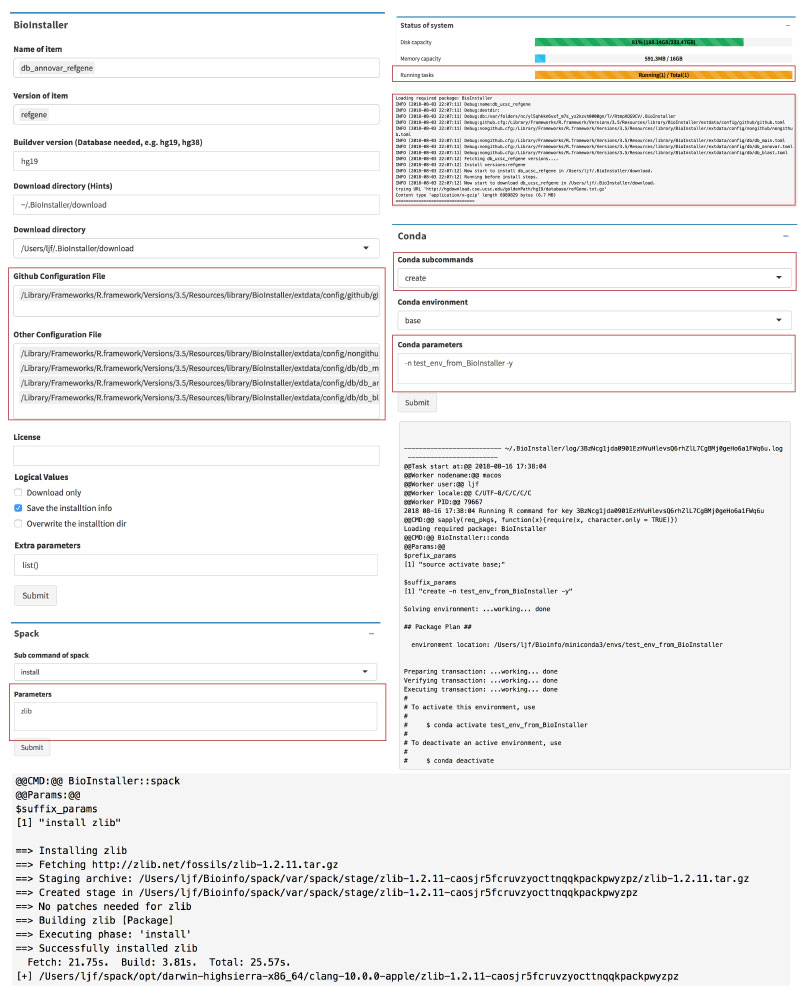
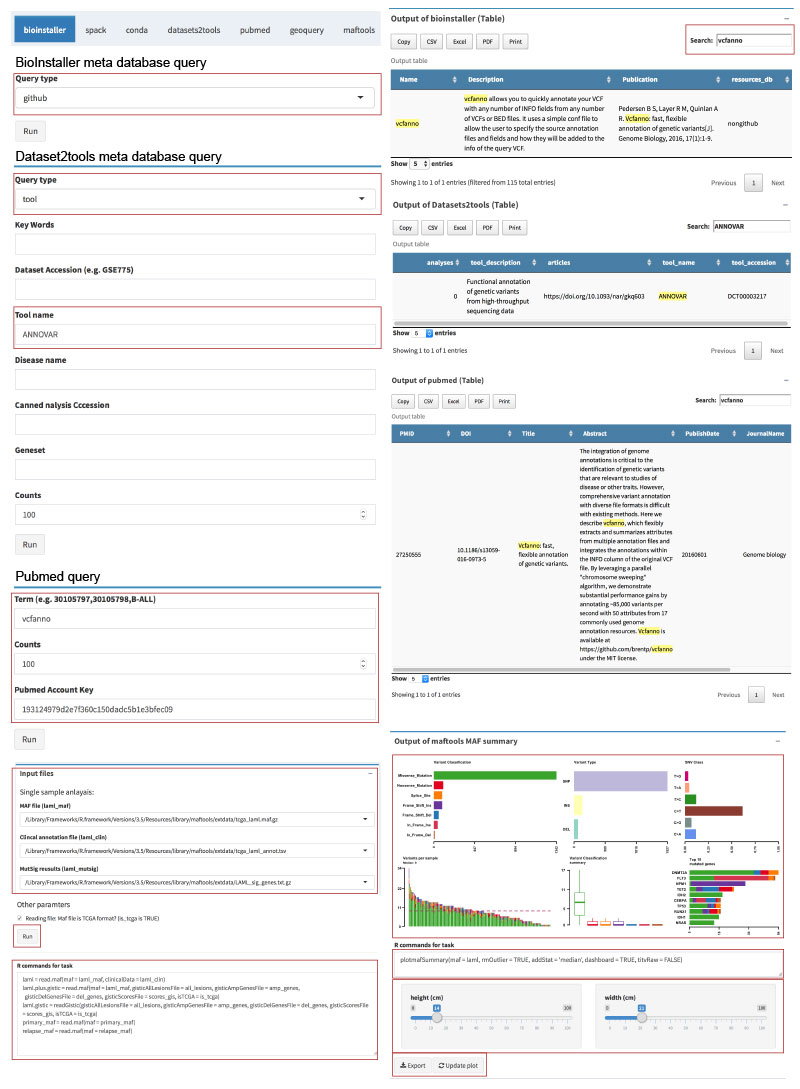 o
o

Installation
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_deps_r
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_start
chmod a+x bioshiny_deps_r
chmod a+x bioshiny_start
# For full toolsets
yarn global add bioshiny
bioshiny_deps_r
Usage
# Set bioshiny main database
echo 'export BIOSHINY_CONFIG="~/.bioshiny/shiny.config.yaml" >> ~/.bashrc'
# Set BioInstaller stored database
echo 'export BIO_SOFTWARES_DB_ACTIVE="~/.bioshiny/info.yaml"'
. ~/.bashrc
Rscript -e "bioshiny::set_shiny_workers(1)"
bioshiny_start
# vcfanno and annovar plugin need to set the tools path
How to contribute?
Please fork the GitHub bioshiny repository, modify it, and submit a pull request to us.
Maintainer
License
MIT