Awesome
pyHH
pyHH is a simple Python implementation of the Hodgkin-Huxley spiking neuron model. pyHH simulates conductances and calculates membrane voltage at discrete time points without requiring a differential equation solver. HHSharp is a similar project written in C#.

Minimal Code Example
A full Hodgkin-Huxley spiking neuron model and simulation was created in fewer than 100 lines of Python (dev/concept4.py). Unlike other code examples on the internet, this implementation is object-oriented and Pythonic. When run, it produces the image above.
Python Package
The pyhh package includes Hodgkin-Huxley models and additional tools to organize simulation data.
Simulation Steps
- Create a model cell and customize its properties if desired
- Create a stimulus waveform (a numpy array)
- Create a simulation by giving it model the waveform you created
- Plot various properties of the stimulation
Example Usage
# customize a neuron model if desired
model = pyhh.HHModel()
model.gNa = 100 # typically 120
model.gK = 5 # typically 36
model.EK = -35 # typically -12
# customize a stimulus waveform
stim = np.zeros(20000)
stim[7000:13000] = 50 # add a square pulse
# simulate the model cell using the custom waveform
sim = pyhh.Simulation(model)
sim.Run(stimulusWaveform=stim, stepSizeMs=0.01)
# plot the results with MatPlotLib
plt.figure(figsize=(10, 8))
ax1 = plt.subplot(411)
ax1.plot(sim.times, sim.Vm - 70, color='b')
ax1.set_ylabel("Potential (mV)")
ax1.set_title("Hodgkin-Huxley Spiking Neuron Model", fontSize=16)
ax2 = plt.subplot(412)
ax2.plot(sim.times, stim, color='r')
ax2.set_ylabel("Stimulation (µA/cm²)")
ax3 = plt.subplot(413, sharex=ax1)
ax3.plot(sim.times, sim.StateH, label='h')
ax3.plot(sim.times, sim.StateM, label='m')
ax3.plot(sim.times, sim.StateN, label='n')
ax3.set_ylabel("Activation (frac)")
ax3.legend()
ax4 = plt.subplot(414, sharex=ax1)
ax4.plot(sim.times, sim.INa, label='VGSC')
ax4.plot(sim.times, sim.IK, label='VGKC')
ax4.plot(sim.times, sim.IKleak, label='KLeak')
ax4.set_ylabel("Current (µA/cm²)")
ax4.set_xlabel("Simulation Time (milliseconds)")
ax4.legend()
plt.tight_layout()
plt.savefig("tests/demo.png")
plt.show()

Theory
Visit https://github.com/swharden/HHSharp for code concepts and simulation notes. Although the language is different, the biology and implementation is the same.
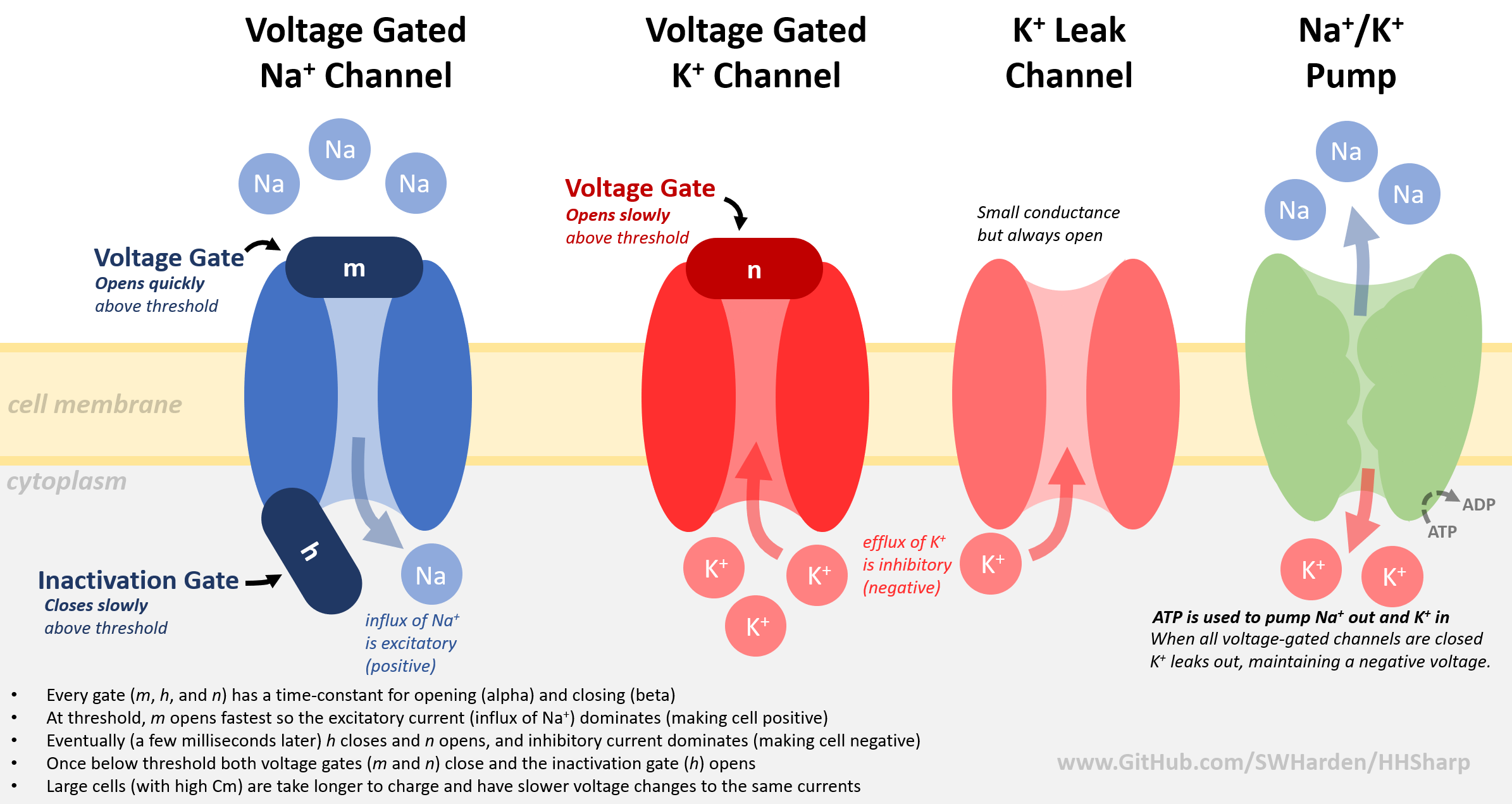
Additional Resources
- Hodgkin and Huxley, 1952 (the original manuscript)
- The Hodgkin-Huxley Mode (The GENESIS Simulator, Chapter 4)
- Wikipedia: Hodgkin–Huxley model
- Hodgkin-Huxley spiking neuron model in Python by Giuseppe Bonaccorso - a HH model which uses the
scipy.integrate.odeintordinary differential equation solver - Introduction to Computational Modeling: Hodgkin-Huxley Model by Andrew Jahn - a commentary of the HH model with matlab code which discretely simulates conductances
- NeuroML Hodgkin Huxley Tutorials
- Summary of the Hodgkin-Huxley model by Dave Beeman