Awesome
MD.ai Python Client Library
Currently pre-alpha -- API may change significantly in future releases.
The python client library is designed to work with the datasets and annotations generated by the MD.ai Medical AI platform.
You can download datasets consisting of images and annotations (i.e., JSON file), create train/validation/test datasets, integrate with various machine learing libraries (e.g., TensorFlow/Keras, Fast.ai) for developing machine learning algorithms.
To get started, check out the examples in the notebooks section, or our intro to deep learning for medical imaging lessons.
Installation
Requires Python 3.6+. Install and update using pip:
pip install --upgrade mdai
Documentation
Overview Documentation: https://docs.md.ai/libraries/python/ API Documentation coming soon.
The MD.ai Annotator
The MD.ai annotator is a powerful web based application, to store and view anonymized medical images (e.g, DICOM) on the cloud, create annotations collaboratively, in real-time, and export annotations, images and labels for training. The MD.ai python client library can be used to download images and annotations, prepare the datasets, and then used to train and evaluate deep learning models.
- MD.ai Documentation and Videos URL: https://docs.md.ai/
- MD.ai Annotator Example Project URL: https://public.md.ai/annotator/project/aGq4k6NW/workspace
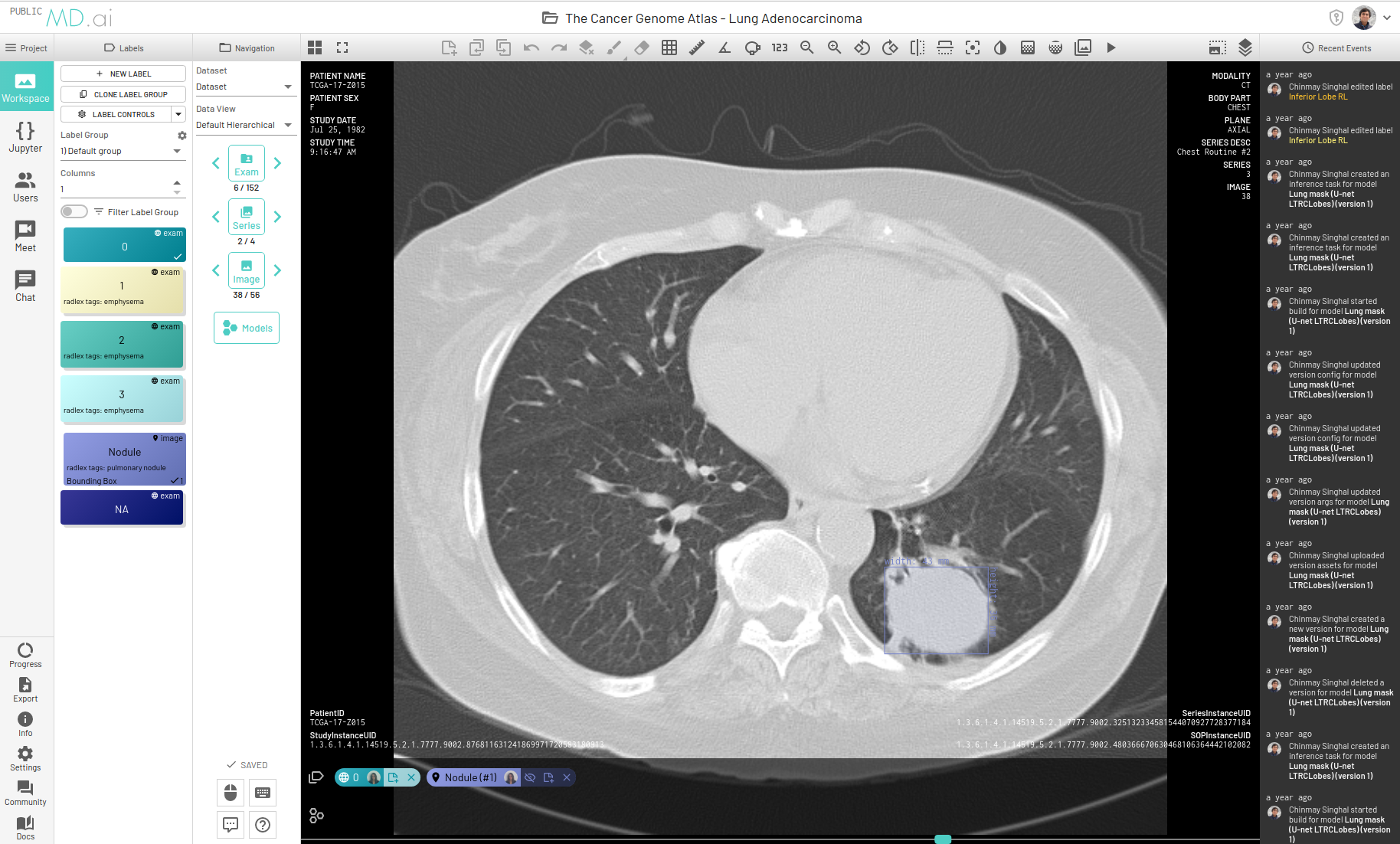
MD.ai Annotation JSON Format
More detailed information regarding the annotation JSON export format, see: https://docs.md.ai/data/json/
Example Notebooks
Introductory lessons to Deep Learning for medical imaging by MD.ai
The following are several Jupyter notebooks covering the basics of downloading and parsing annotation data, and training and evaluating different deep learning models for classification, semantic and instance segmentation and object detection problems in the medical imaging domain. The notebooks can be run on Google's colab with GPU (see instruction below).
- Lesson 1. Classification of chest vs. adominal X-rays using TensorFlow/Keras Github | Annotator
- Lesson 2. Lung X-Rays Semantic Segmentation using UNets. Github | Annotator
- Lesson 3. RSNA Pneumonia detection using Kaggle data format Github | Annotator
- Lesson 3. RSNA Pneumonia detection using MD.ai python client library Github | Annotator
Contributing
See contributing guidelines to set up a development environemnt and how to make contributions to mdai.
Running Jupyter notebooks Colab
It’s easy to run a Jupyter notebook on Google's Colab with free GPU use (time limited). For example, you can add the Github jupyter notebook path to https://colab.research.google.com/notebook: Select the "GITHUB" tab, and add the Lesson 1 URL: https://github.com/mdai/ml-lessons/blob/master/lesson1-xray-images-classification.ipynb
To use the GPU, in the notebook menu, go to Runtime -> Change runtime type -> switch to Python 3, and turn on GPU. See more colab tips.
Advanced: How to run on Google Cloud Platform with Deep Learning Images
You can also run the notebook with powerful GPUs on the Google Cloud Platform. In this case, you need to authenticate to the Google Cloug Platform, create a private virtual machine instance running a Google's Deep Learning image, and import the lessons. See instructions below.
GCP Deep Learnings Images How To
© 2023 MD.ai, Inc.