Awesome
testing-ml
How to test machine learning code. In this example, we'll test a numpy implementation of DecisionTree and RandomForest via:
- Pre-train tests to ensure correct implementation
- Post-train tests to ensure expected learned behaviour
- Evaluation to ensure satisfactory model performance
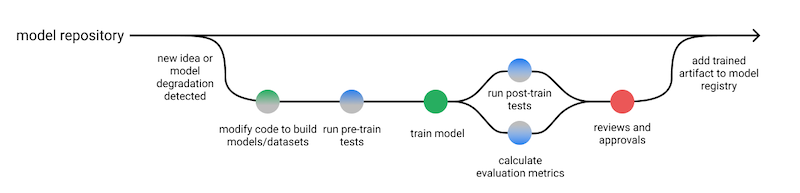
Accompanying article: How to Test Machine Learning Code and Systems. Inspired by @jeremyjordan's Effective Testing for Machine Learning Systems.
Quick Start
# Clone and setup environment
git clone https://github.com/eugeneyan/testing-ml.git
cd testing-ml
make setup
# Run test suite
make check
Standard software habits
- Unit test fixture reuse, exceptions testing with
pytest - Code coverage with
Coverage.pyandpytest-cov - Linting to ensure code consistency with
pylint - Type checks to verify type correctness with
mypy
More details here: How to Set Up a Python Project For Automation and Collaboration (GitHub repo)
Pre-train tests to ensure correct implementation
- Test implementation of Gini impurity and gain
def test_gini_impurity():
assert round(gini_impurity([1, 1, 1, 1, 1, 1, 1, 1]), 3) == 0
assert round(gini_impurity([1, 1, 1, 1, 1, 1, 0, 0]), 3) == 0.375
assert round(gini_impurity([1, 1, 1, 1, 0, 0, 0, 0]), 3) == 0.500
def test_gini_gain():
assert round(gini_gain([1, 1, 1, 1, 0, 0, 0, 0], [[1, 1, 1, 1], [0, 0, 0, 0]]), 3) == 0.5
assert round(gini_gain([1, 1, 1, 1, 0, 0, 0, 0], [[1, 1, 1, 0], [0, 0, 0, 1]]), 3) == 0.125
assert round(gini_gain([1, 1, 1, 1, 0, 0, 0, 0], [[1, 1, 0, 0], [0, 0, 1, 1]]), 3) == 0.0
- Test output shape
def test_dt_output_shape(dummy_titanic):
X_train, y_train, X_test, y_test = dummy_titanic
dt = DecisionTree()
dt.fit(X_train, y_train)
pred_train, pred_test = dt.predict(X_train), dt.predict(X_test)
assert pred_train.shape == (X_train.shape[0],), 'DecisionTree output should be same as training labels.'
assert pred_test.shape == (X_test.shape[0],), 'DecisionTree output should be same as testing labels.'
- Test data leak between train and test set
def test_data_leak_in_test_data(dummy_titanic_df):
train, test = dummy_titanic_df
concat_df = pd.concat([train, test])
concat_df.drop_duplicates(inplace=True)
assert concat_df.shape[0] == train.shape[0] + test.shape[0]
- Test output range
def test_dt_output_range(dummy_titanic):
X_train, y_train, X_test, y_test = dummy_titanic
dt = DecisionTree()
dt.fit(X_train, y_train)
pred_train, pred_test = dt.predict(X_train), dt.predict(X_test)
assert (pred_train <= 1).all() & (pred_train >= 0).all(), 'Decision tree output should range from 0 to 1 inclusive'
assert (pred_test <= 1).all() & (pred_test >= 0).all(), 'Decision tree output should range from 0 to 1 inclusive'
- Test model able to overfit on perfectly separable data
def test_dt_overfit(dummy_feats_and_labels):
feats, labels = dummy_feats_and_labels
dt = DecisionTree()
dt.fit(feats, labels)
pred = np.round(dt.predict(feats))
assert np.array_equal(labels, pred), 'DecisionTree should fit data perfectly and prediction should == labels.'
- Test additional tree depth increases training accuracy and AUC ROC
def test_dt_increase_acc(dummy_titanic):
X_train, y_train, _, _ = dummy_titanic
acc_list, auc_list = [], []
for depth in range(1, 10):
dt = DecisionTree(depth_limit=depth)
dt.fit(X_train, y_train)
pred = dt.predict(X_train)
pred_binary = np.round(pred)
acc_list.append(accuracy_score(y_train, pred_binary))
auc_list.append(roc_auc_score(y_train, pred))
assert sorted(acc_list) == acc_list, 'Accuracy should increase as tree depth increases.'
assert sorted(auc_list) == auc_list, 'AUC ROC should increase as tree depth increases.'
- Test additional trees in
RandomForestimproves validation accuracy and AUC ROC
def test_dt_increase_acc(dummy_titanic):
X_train, y_train, X_test, y_test = dummy_titanic
acc_list, auc_list = [], []
for num_trees in [1, 3, 7, 15]:
rf = RandomForest(num_trees=num_trees, depth_limit=7, col_subsampling=0.7, row_subsampling=0.7)
rf.fit(X_train, y_train)
pred = rf.predict(X_test)
pred_binary = np.round(pred)
acc_list.append(accuracy_score(y_test, pred_binary))
auc_list.append(roc_auc_score(y_test, pred))
assert sorted(acc_list) == acc_list, 'Accuracy should increase as number of trees increases.'
assert sorted(auc_list) == auc_list, 'AUC ROC should increase as number of trees increases.'
- Test
RandomForestoutperformsDecisionTreegiven the same tree depth
def test_rf_better_than_dt(dummy_titanic):
X_train, y_train, X_test, y_test = dummy_titanic
dt = DecisionTree(depth_limit=10)
dt.fit(X_train, y_train)
rf = RandomForest(depth_limit=10, num_trees=7, col_subsampling=0.8, row_subsampling=0.8)
rf.fit(X_train, y_train)
pred_test_dt = dt.predict(X_test)
pred_test_binary_dt = np.round(pred_test_dt)
acc_test_dt = accuracy_score(y_test, pred_test_binary_dt)
auc_test_dt = roc_auc_score(y_test, pred_test_dt)
pred_test_rf = rf.predict(X_test)
pred_test_binary_rf = np.round(pred_test_rf)
acc_test_rf = accuracy_score(y_test, pred_test_binary_rf)
auc_test_rf = roc_auc_score(y_test, pred_test_rf)
assert acc_test_rf > acc_test_dt, 'RandomForest should have higher accuracy than DecisionTree on test set.'
assert auc_test_rf > auc_test_dt, 'RandomForest should have higher AUC ROC than DecisionTree on test set.'
Post-train tests to ensure expected learned behaviour
- Test invariance (e.g., ticket number should not affect survival probability)
def test_dt_invariance(dummy_titanic_dt, dummy_passengers):
model = dummy_titanic_dt
_, p2 = dummy_passengers
# Get original survival probability of passenger 2
test_df = pd.DataFrame.from_dict([p2], orient='columns')
X, y = get_feats_and_labels(prep_df(test_df))
p2_prob = model.predict(X)[0] # 1.0
# Change ticket number from 'PC 17599' to 'A/5 21171'
p2_ticket = p2.copy()
p2_ticket['ticket'] = 'A/5 21171'
test_df = pd.DataFrame.from_dict([p2_ticket], orient='columns')
X, y = get_feats_and_labels(prep_df(test_df))
p2_ticket_prob = model.predict(X)[0] # 1.0
assert p2_prob == p2_ticket_prob
- Test directional expectation (e.g., females should have higher survival probability than males)
def test_dt_directional_expectation(dummy_titanic_dt, dummy_passengers):
model = dummy_titanic_dt
_, p2 = dummy_passengers
# Get original survival probability of passenger 2
test_df = pd.DataFrame.from_dict([p2], orient='columns')
X, y = get_feats_and_labels(prep_df(test_df))
p2_prob = model.predict(X)[0] # 1.0
# Change gender from female to male
p2_male = p2.copy()
p2_male['Name'] = ' Mr. John'
p2_male['Sex'] = 'male'
test_df = pd.DataFrame.from_dict([p2_male], orient='columns')
X, y = get_feats_and_labels(prep_df(test_df))
p2_male_prob = model.predict(X)[0] # 0.56
# Change class from 1 to 3
p2_class = p2.copy()
p2_class['Pclass'] = 3
test_df = pd.DataFrame.from_dict([p2_class], orient='columns')
X, y = get_feats_and_labels(prep_df(test_df))
p2_class_prob = model.predict(X)[0] # 0.0
assert p2_prob > p2_male_prob, 'Changing gender from female to male should decrease survival probability.'
assert p2_prob > p2_class_prob, 'Changing class from 1 to 3 should decrease survival probability.'
Evaluation to ensure satisfactory model performance
- Evaluation on accuracy and AUC ROC
def test_dt_evaluation(dummy_titanic_dt, dummy_titanic):
model = dummy_titanic_dt
X_train, y_train, X_test, y_test = dummy_titanic
pred_test = model.predict(X_test)
pred_test_binary = np.round(pred_test)
acc_test = accuracy_score(y_test, pred_test_binary)
auc_test = roc_auc_score(y_test, pred_test)
assert acc_test > 0.82, 'Accuracy on test should be > 0.82'
assert auc_test > 0.84, 'AUC ROC on test should be > 0.84'
def test_dt_training_time(dummy_titanic):
X_train, y_train, X_test, y_test = dummy_titanic
# Standardize to use depth = 10
dt = DecisionTree(depth_limit=10)
latency_array = np.array([train_with_time(dt, X_train, y_train)[1] for i in range(100)])
time_p95 = np.quantile(latency_array, 0.95)
assert time_p95 < 1.0, 'Training time at 95th percentile should be < 1.0 sec'
def test_dt_serving_latency(dummy_titanic):
X_train, y_train, X_test, y_test = dummy_titanic
# Standardize to use depth = 10
dt = DecisionTree(depth_limit=10)
dt.fit(X_train, y_train)
latency_array = np.array([predict_with_time(dt, X_test)[1] for i in range(500)])
latency_p99 = np.quantile(latency_array, 0.99)
assert latency_p99 < 0.004, 'Serving latency at 99th percentile should be < 0.004 sec'
