Awesome
<!-- README.md is generated from README.Rmd. Please edit that file -->easyalluvial <a href='https://erblast.github.io/easyalluvial'><img src='man/figures/logo.png' align="right" height="139" /></a>
Alluvial plots are similar to sankey
diagrams and visualise
categorical data over multiple dimensions as flows. Rosval et.
al. 2010
Their graphical grammar however is a bit more complex then that of a
regular x/y plots. The
ggalluvial package made a
great job of translating that grammar into
ggplot2 syntax and gives you
many option to tweak the appearance of an alluvial plot, however there
still remains a multi-layered complexity that makes it difficult to use
‘ggalluvial’ for explorative data analysis. ‘easyalluvial’ provides a
simple interface to this package that allows you to produce a decent
alluvial plot from any dataframe in either long or wide format from a
single line of code while also handling continuous data. It is meant to
allow a quick visualisation of entire dataframes with a focus on
different colouring options that can make alluvial plots a great tool
for data exploration.
Features
- plot alluvial graph with a single line of code of a given dataframe
- support for wide and long data format (wiki, wide vs. long/narrow data)
- automatically transforms numerical to categorical data
- helper functions for variable selection
- convenient parameters for coloring and ordering
- marginal histograms
- model agnostic partial dependence and model response alluvial plots with 4 dimensions
- interactive plots with
easyalluvialandparcats
Installation
CRAN
install.packages('easyalluvial')
Development Version
# install.packages("devtools")
devtools::install_github("erblast/easyalluvial")
Documentation
Examples
suppressPackageStartupMessages( require(tidyverse) )
suppressPackageStartupMessages( require(easyalluvial) )
Alluvial from data in wide format
Sample Data
knitr::kable( head(mtcars2) )
| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb | ids |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | V | manual | 4 | 4 | Mazda RX4 |
| 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | V | manual | 4 | 4 | Mazda RX4 Wag |
| 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | S | manual | 4 | 1 | Datsun 710 |
| 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | S | automatic | 3 | 1 | Hornet 4 Drive |
| 18.7 | 8 | 360 | 175 | 3.15 | 3.440 | 17.02 | V | automatic | 3 | 2 | Hornet Sportabout |
| 18.1 | 6 | 225 | 105 | 2.76 | 3.460 | 20.22 | S | automatic | 3 | 1 | Valiant |
Plot
Continuous Variables will be automatically binned as follows.
- High, High (HH)
- Medium, High (MH)
- Medium (M)
- Medium, Low (ML)
- Low, Low (LL)
alluvial_wide( data = mtcars2
, max_variables = 5
, fill_by = 'first_variable' )
 <!-- -->
<!-- -->
Alluvial from data in long format
Sample Data
knitr::kable( head(quarterly_flights) )
| tailnum | carrier | origin | dest | qu | mean_arr_delay |
|---|---|---|---|---|---|
| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q1 | on_time |
| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q2 | on_time |
| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q3 | on_time |
| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q4 | on_time |
| N11150 EWR MCI EV | EV | EWR | MCI | Q1 | late |
| N11150 EWR MCI EV | EV | EWR | MCI | Q2 | late |
Plot
alluvial_long( quarterly_flights
, key = qu
, value = mean_arr_delay
, id = tailnum
, fill = carrier )
 <!-- -->
<!-- -->
Marginal Histograms
alluvial_wide( data = mtcars2
, max_variables = 5
, fill_by = 'first_variable' ) %>%
add_marginal_histograms(mtcars2)
 <!-- -->
<!-- -->
Interactive Graphs
suppressPackageStartupMessages( require(parcats) )
p = alluvial_wide(mtcars2, max_variables = 5)
parcats(p, marginal_histograms = TRUE, data_input = mtcars2)
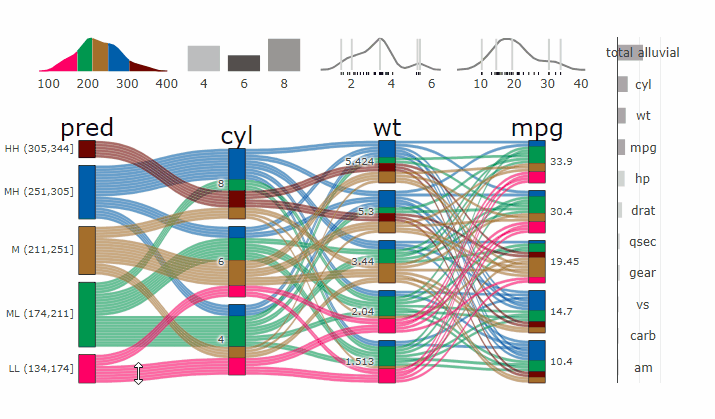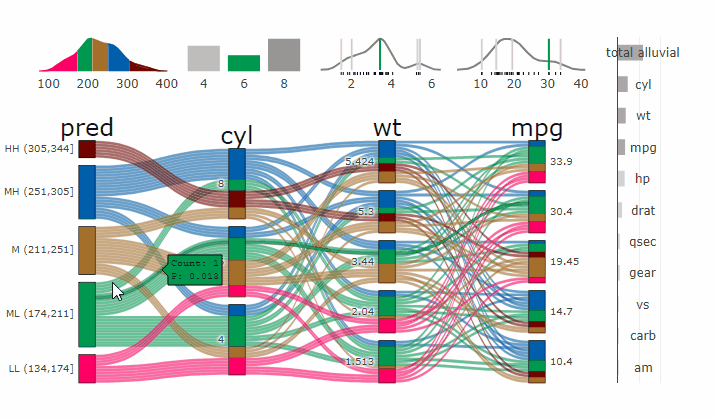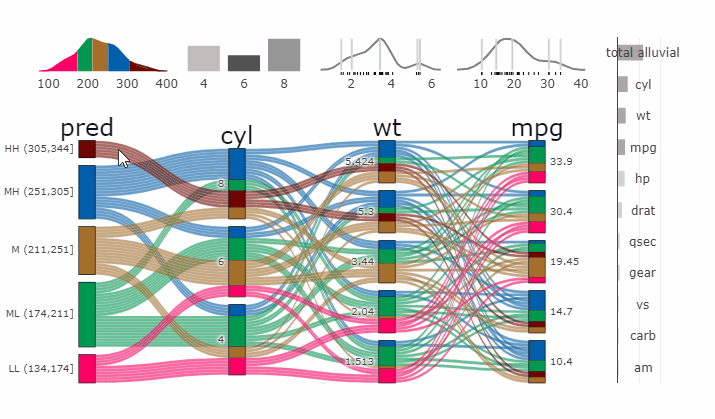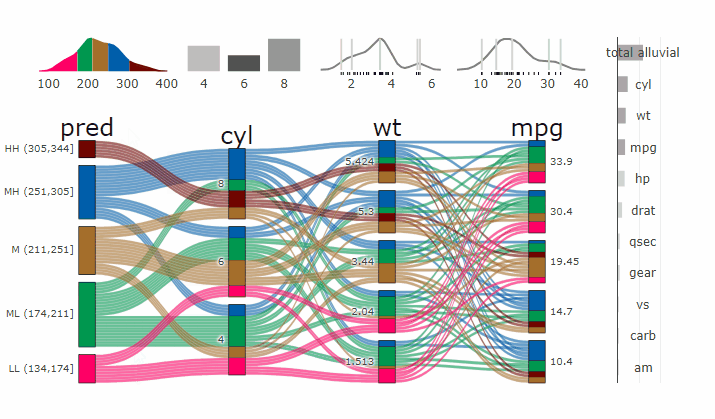
Partial Dependence Alluvial Plots
Alluvial plots are capable of displaying higher dimensional data on a plane, thus lend themselves to plot the response of a statistical model to changes in the input data across multiple dimensions. The practical limit here is 4 dimensions while conventional partial dependence plots are limited to 2 dimensions.
Briefly the 4 variables with the highest feature importance for a given model are selected and 5 values spread over the variable range are selected for each. Then a grid of all possible combinations is created. All none-plotted variables are set to the values found in the first row of the training data set. Using this artificial data space model predictions are being generated. This process is then repeated for each row in the training data set and the overall model response is averaged in the end. Each of the possible combinations is plotted as a flow which is coloured by the bin corresponding to the average model response generated by that particular combination.
easyalluvial contains wrappers for parsnip and caret models.
Custom Wrappers for other models can easily be created.
df = select(mtcars2, -ids)
m = parsnip::rand_forest(mode = "regression") %>%
parsnip::set_engine("randomForest") %>%
parsnip::fit(disp ~ ., df)
p = alluvial_model_response_parsnip(m, df, degree = 4, method = "pdp")
#> Getting partial dependence plot preditions. This can take a while. See easyalluvial::get_pdp_predictions() `Details` on how to use multiprocessing
p_grid = add_marginal_histograms(p, df, plot = F) %>%
add_imp_plot(p, df)
 <!-- -->
<!-- -->
Interactive Partial Dependence Plot
parcats(p, marginal_histograms = TRUE, imp = TRUE, data_input = df)
ClinicoPath {jamovi} Module
ClinicoPath jamovi
Module (thanks to
Serdar Balci) adds easyalluvial plots to jamovia spreadsheet
interface for doing statistics with R.

 -
-