Awesome
alevinQC
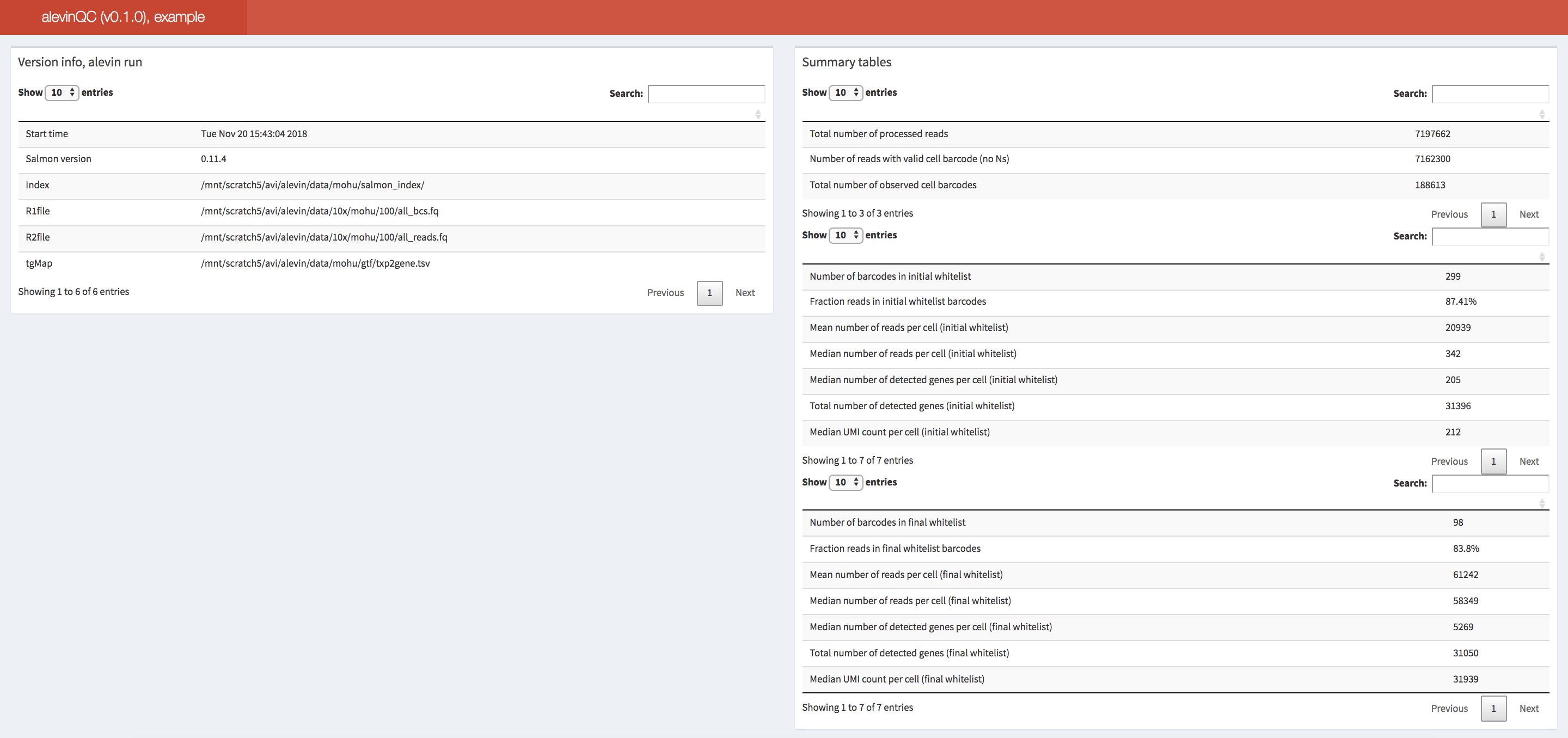
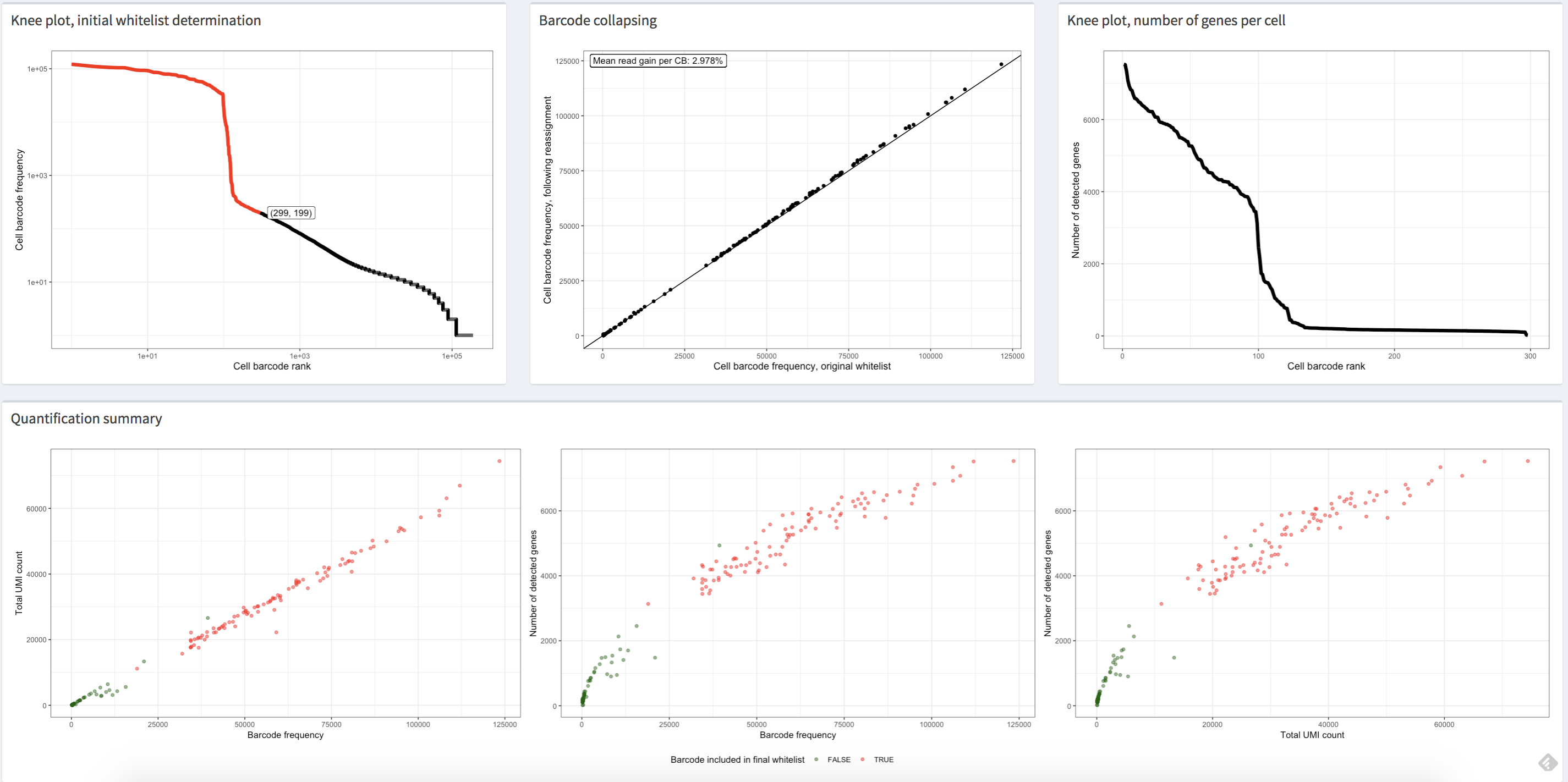
The alevinQC R package provides functionality for generating QC reports
summarizing the output of alevin
(Srivastava et al., Genome Biology 20:65 (2019)). The reports can
be generated in html or pdf format, or as R/Shiny applications.
Installation:
alevinQC is available from
Bioconductor, and can be
installed using the BiocManager CRAN package:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("alevinQC")
Note that alevinQC v 1.1 or newer is required to process output from Salmon version 0.14.0 or newer.
Example usage:
alevinQCReport(baseDir = system.file("extdata/alevin_example_v0.14",
package = "alevinQC"),
sampleId = "testSample",
outputFile = "alevinReport.html",
outputFormat = "html_document",
outputDir = tempdir(), forceOverwrite = TRUE)
For more information, we refer to the package vignette.