Awesome
MaskingDepth (IROS 2024)
This code is the implementation of the paper <a href="https://arxiv.org/abs/2212.10806">MaskingDepth: Masked Consistency Regularization for Semi-supervised Monocular Depth Estimation</a> by Baek et al.
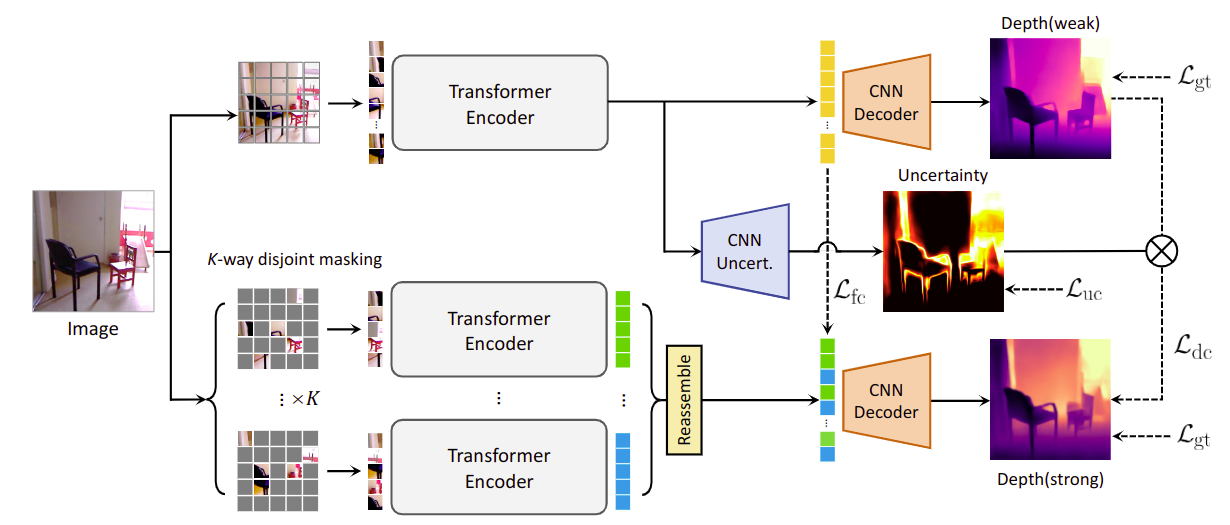
We propose MaskingDepth, a novel semi-supervised learning framework for monocular depth estimation to mitigate the reliance on large ground-truth depth quantities. MaskingDepth is designed to enforce consistency between the strongly-augmented unlabeled data and the pseudo-labels derived from weakly-augmented unlabeled data, which enables learning depth without supervision. In this framework, a novel data augmentation is proposed to take the advantage of a naive masking strategy as an augmentation, while avoiding its scale ambiguity problem between depths from weakly- and strongly-augmented branches and risk of missing small-scale instances. To only retain high-confident depth predictions from the weakly-augmented branch as pseudo-labels, we also present an uncertainty estimation technique, which is used to define robust consistency regularization. Experiments on KITTI and NYU-Depth-v2 datasets demonstrate the effectiveness of each component, its robustness to the use of fewer depth-annotated images, and superior performance compared to other state-of-the-art semi-supervised methods for monocular depth estimation.
Environment
- NGC pytorch 20.11-py3 (Docker container)
- additional require packages (dotmap, wandb, einops, timm)
- NVIDIA RGX 3090s
In docker container
git clone https://github.com/KU-CVLAB/MaskingDepth.git # Download this project
cd MaskingDepth # Change directory
sh package_install.sh # Install additionally package
we recommend that vit encoder initialize through this weight file. Download ViT ImageNet pretrained weight
Dataset
Training
- Supervised learning:
Edit conf/base_train.yaml file. See the comments in the configuration file for detail options.
python train.py
- Semi-supervised learning
Edit conf/consistency_train.yaml file. See the comments in the configuration file for detail options.
python consistency_train.py
Evaluation
We evaluate through the eval_with_pngs.py created by BTS. For evaluation we divide test set according to Eigen split.
Results
Quantitative results on the KITTI dataset in a sparsely-supervised setting
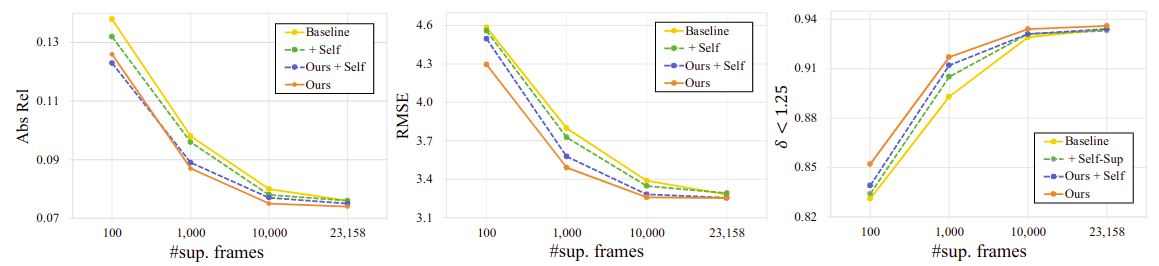
full
| Methods | AbsRel ↓ | SqRel ↓ | RMSE ↓ | RMSElog ↓ | δ↑ |
|---|---|---|---|---|---|
| Baseline | 0.076 ± 0.003 | 0.365 ± 0.004 | 3.290 ± 0.015 | 0.118 ± 0.001 | 0.934 ± 0.001 |
| Baseline+Self | 0.076 ± 0.002 | 0.367 ± 0.007 | 3.291 ± 0.020 | 0.117 ± 0.001 | 0.933 ± 0.002 |
| Ours+Self | 0.079 ± 0.001 | 0.379 ± 0.007 | 3.388 ± 0.019 | 0.121 ± 0.009 | 0.929 ± 0.001 |
| Ours | 0.074 ± 0.001 | 0.362 ± 0.001 | 3.253 ± 0.012 | 0.116 ± 0.001 | 0.935 ± 0.001 |
10,000
| Methods | AbsRel ↓ | SqRel ↓ | RMSE ↓ | RMSElog ↓ | δ↑ |
|---|---|---|---|---|---|
| Baseline | 0.079 ± 0.001 | 0.379 ± 0.007 | 3.388 ± 0.019 | 0.121 ± 0.009 | 0.929 ± 0.001 |
| Baseline+Self | 0.078 ± 0.001 | 0.376 ± 0.006 | 3.347 ± 0.043 | 0.119 ± 0.002 | 0.931 ± 0.001 |
| Ours+Self | 0.076 ± 0.017 | 0.369 ± 0.004 | 3.311 ± 0.011 | 0.117 ± 0.001 | 0.935 ± 0.002 |
| Ours | 0.075 ± 0.002 | 0.362 ± 0.006 | 3.259 ± 0.020 | 0.116 ± 0.001 | 0.934 ± 0.003 |
1,000
| Methods | AbsRel ↓ | SqRel ↓ | RMSE ↓ | RMSElog ↓ | δ↑ |
|---|---|---|---|---|---|
| Baseline | 0.098 ± 0.004 | 0.515 ± 0.030 | 3.785 ± 0.013 | 0.142 ± 0.005 | 0.899 ± 0.005 |
| Baseline+Self | 0.096 ± 0.002 | 0.523 ± 0.024 | 3.750 ± 0.033 | 0.140 ± 0.002 | 0.900 ± 0.004 |
| Ours+Self | 0.085 ± 0.017 | 0.430 ± 0.011 | 3.521 ± 0.012 | 0.129 ± 0.012 | 0.918 ± 0.010 |
| Ours | 0.088 ± 0.003 | 0.419 ± 0.007 | 3.490 ± 0.020 | 0.129 ± 0.003 | 0.917 ± 0.002 |
100
| Methods | AbsRel ↓ | SqRel ↓ | RMSE ↓ | RMSElog ↓ | δ↑ |
|---|---|---|---|---|---|
| Baseline | 0.135 ± 0.005 | 0.728 ± 0.019 | 4.585 ± 0.048 | 0.186 ± 0.011 | 0.831 ± 0.005 |
| Baseline+Self | 0.132 ± 0.004 | 0.759 ± 0.014 | 4.559 ± 0.044 | 0.184 ± 0.003 | 0.834 ± 0.004 |
| Ours+Self | 0.123 ± 0.003 | 0.747 ± 0.018 | 4.497 ± 0.042 | 0.181 ± 0.005 | 0.839 ± 0.005 |
| Ours | 0.128 ± 0.004 | 0.707 ± 0.013 | 4.295 ± 0.037 | 0.173 ± 0.006 | 0.849 ± 0.006 |
10
| Methods | AbsRel ↓ | SqRel ↓ | RMSE ↓ | RMSElog ↓ | δ↑ |
|---|---|---|---|---|---|
| Baseline | 0.201 ± 0.023 | 1.508 ± 0.045 | 6.163 ± 0.082 | 0.268 ± 0.029 | 0.701 ± 0.021 |
| Baseline+Self | 0.210 ± 0.020 | 1.322 ± 0.042 | 5.627 ± 0.080 | 0.265 ± 0.027 | 0.711 ± 0.016 |
| Ours+Self | 0.184 ± 0.011 | 1.265 ± 0.064 | 5.747 ± 0.080 | 0.243 ± 0.007 | 0.727 ± 0.018 |
| Ours | 0.197 ± 0.019 | 1.378 ± 0.032 | 5.650 ± 0.091 | 0.261 ± 0.030 | 0.723 ± 0.017 |
Quantitative results on the NYU-Depth-v2 dataset in a sparsely-supervised setting
full
| Methods | AbsRel ↓ | RMSE ↓ | log10 ↓ | δ↑ |
|---|---|---|---|---|
| Baseline | 0.106 ± 0.002 | 0.380 ± 0.004 | 0.053 ± 0.001 | 0.897 ± 0.001 |
| Ours | 0.105 ± 0.002 | 0.379 ± 0.003 | 0.053 ± 0.001 | 0.899 ± 0.001 |
10,000
| Methods | AbsRel ↓ | RMSE ↓ | log10 ↓ | δ↑ |
|---|---|---|---|---|
| Baseline | 0.112 ± 0.004 | 0.389 ± 0.006 | 0.057 ± 0.003 | 0.893 ± 0.003 |
| Ours | 0.107 ± 0.002 | 0.386 ± 0.006 | 0.054 ± 0.002 | 0.896 ± 0.002 |
1,000
| Methods | AbsRel ↓ | RMSE ↓ | log10 ↓ | δ↑ |
|---|---|---|---|---|
| Baseline | 0.141 ± 0.008 | 0.447 ± 0.009 | 0.066 ± 0.004 | 0.843 ± 0.006 |
| Ours | 0.135 ± 0.007 | 0.440 ± 0.008 | 0.065 ± 0.004 | 0.853 ± 0.005 |
100
| Methods | AbsRel ↓ | RMSE ↓ | log10 ↓ | δ↑ |
|---|---|---|---|---|
| Baseline | 0.199 ± 0.011 | 0.604 ± 0.014 | 0.086 ± 0.005 | 0.694 ± 0.011 |
| Ours | 0.182 ± 0.008 | 0.594 ± 0.012 | 0.083 ± 0.003 | 0.718 ± 0.010 |
10
| Methods | AbsRel ↓ | RMSE ↓ | log10 ↓ | δ↑ |
|---|---|---|---|---|
| Baseline | 0.321 ± 0.040 | 0.872 ± 0.042 | 0.124 ± 0.008 | 0.523 ± 0.027 |
| Ours | 0.292 ± 0.031 | 0.814 ± 0.037 | 0.112 ± 0.006 | 0.561 ± 0.021 |
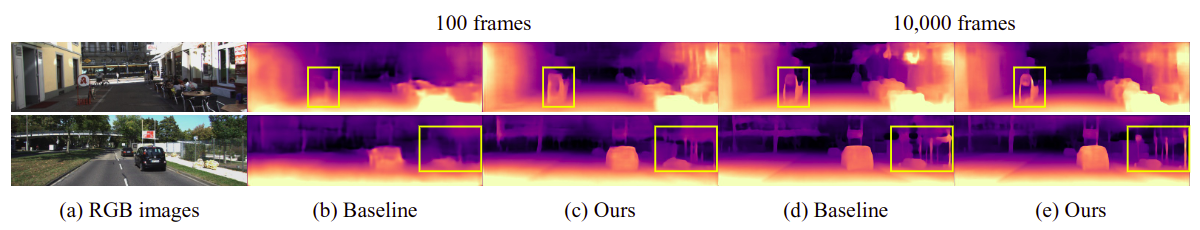
Qualitative results on the KITTI dataset. (a) RGB image, predicted depth maps by (b), (d) baseline, and (c), (e) ours using 100 and 10,000 labeled frames, respectively.
Qualitative results on the NYU-Depth-v2 dataset. (a) RGB image, (b) ground-truth depth map, and predicted depth maps by (c), (e) baseline, and (d), (f) ours using 100 and 10,000 labeled frames, respectively.

Citation
Please consider citing our paper if you use this code.
@article{baek2022semi,
title={Semi-Supervised Learning of Monocular Depth Estimation via Consistency Regularization with K-way Disjoint Masking},
author={Baek, Jongbeom and Kim, Gyeongnyeon and Park, Seonghoon and An, Honggyu and Poggi, Matteo and Kim, Seungryong},
journal={arXiv preprint arXiv:2212.10806},
year={2022}
}