Awesome
MedAGI: Unify Domain-specific Medical LLMs with the Lowest Cost
Juexiao Zhou#, Xiuying Chen#, Xin Gao. #Equal Contribution
King Abdullah University of Science and Technology, KAUST
<a href='MedAGI_v1.pdf'><img src='https://img.shields.io/badge/Paper-PDF-red'></a>
Installation
conda env create -f environment.yml
conda activate medagi
Preparation of Domain-specific Alignment layers
To train your domain-specific layer, please follow the steps at https://github.com/Vision-CAIR/MiniGPT-4
First, set the path to vicuna in medagi/configs/models/medagi.yaml
Then set the model path (ckpt_list) and model description (model_description_list) in eval_configs/medagi_eval.yaml for each of your domain-specific alignment layers, for example
ckpt_list: ['MedAGI/weights/skingpt_v4.2.pth',
'MedAGI/weights/xray.pth',
'MedAGI/weights/pathology.pth',
]
model_description_list: [
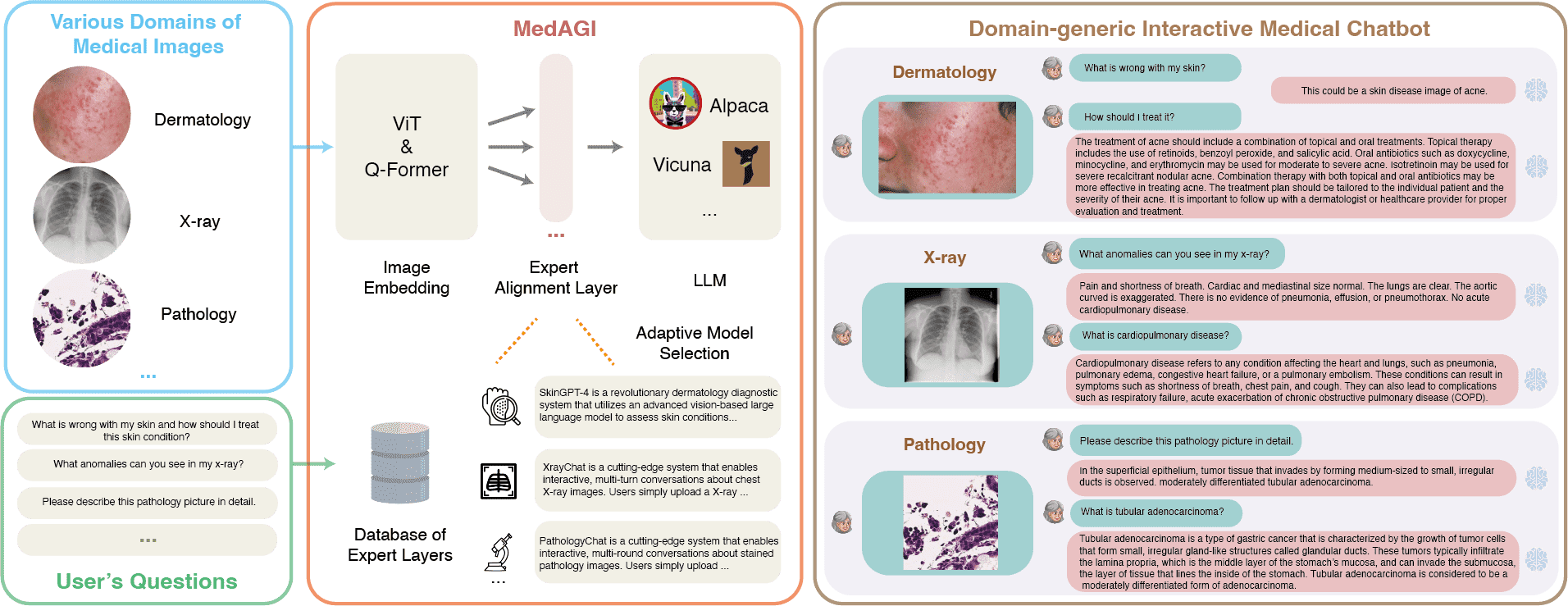
'SkinGPT is a revolutionary dermatology diagnostic system that utilizes an advanced vision-based large language model to assess skin conditions. By uploading personal skin photos to the system, users receive an autonomous analysis that can identify and categorize various skin conditions, and provide treatment recommendations.',
'XrayChat is a cutting-edge system that enables interactive, multi-turn conversations about chest X-ray images. Users simply upload a chest X-ray image, ask any question about it, and XrayChat generates informed responses. The system utilizes an X-ray encoder, a large language model, and an adaptor to comprehend the X-ray image and produce accurate and helpful answers.',
'PathologyChat is a cutting-edge system that enables interactive, multi-round conversations about stained pathology images. Users simply upload a pathology, ask any question about it, and PathologyChat generates informed responses.'
]
Now, you are ready to go!
MedAGI will pick up the most suitable domain-specific alignment layer for you according to your inputs.
Launching Demo Locally
python demo.py --cfg-path eval_configs/medagi_eval.yaml --gpu-id 0
To Contribute
We are open to accepting more domain-specific alignment layers! Please PR to add your domain-specific alignment layer and description in eval_configs/medagi_eval.yaml and place your model under ./weights.
Acknowledgement
- MiniGPT-4 This repo is based on MiniGPT-4, an awesome repo for vision-language chatbot!
- Lavis
- Vicuna
Citation
If you're using MedAGI in your research or applications, please cite MedAGI using this BibTeX:
@misc{zhou2023path,
title={Path to Medical AGI: Unify Domain-specific Medical LLMs with the Lowest Cost},
author={Juexiao Zhou and Xiuying Chen and Xin Gao},
year={2023},
eprint={2306.10765},
archivePrefix={arXiv},
primaryClass={cs.AI}
}