Awesome
Exact
This is a collaborative online tool for labeling image data.
Reference
This paper describes the EXACT-Server in depth. Please cite if you use this tool in your research:
Marzahl et al. EXACT: A collaboration toolset for algorithm-aided annotation of almost everything
@Article{marzahl2021exact,
title={EXACT: a collaboration toolset for algorithm-aided annotation of images with annotation version control},
author={Marzahl, Christian and Aubreville, Marc and Bertram, Christof A. and Maier, Jennifer and Bergler, Christian and Kr{\"o}ger, Christine and Voigt, J{\"o}rn and Breininger, Katharina and Klopfleisch, Robert and Maier, Andreas},
journal={Scientific Reports},
year={2021},
month={Feb},
day={23},
volume={11},
number={1},
pages={4343},
abstract={In many research areas, scientific progress is accelerated by multidisciplinary access to image data and their interdisciplinary annotation. However, keeping track of these annotations to ensure a high-quality multi-purpose data set is a challenging and labour intensive task. We developed the open-source online platform EXACT (EXpert Algorithm Collaboration Tool) that enables the collaborative interdisciplinary analysis of images from different domains online and offline. EXACT supports multi-gigapixel medical whole slide images as well as image series with thousands of images. The software utilises a flexible plugin system that can be adapted to diverse applications such as counting mitotic figures with a screening mode, finding false annotations on a novel validation view, or using the latest deep learning image analysis technologies. This is combined with a version control system which makes it possible to keep track of changes in the data sets and, for example, to link the results of deep learning experiments to specific data set versions. EXACT is freely available and has already been successfully applied to a broad range of annotation tasks, including highly diverse applications like deep learning supported cytology scoring, interdisciplinary multi-centre whole slide image tumour annotation, and highly specialised whale sound spectroscopy clustering.},
issn={2045-2322},
doi={10.1038/s41598-021-83827-4},
url={https://doi.org/10.1038/s41598-021-83827-4}
}
Features
- Browsable REST-API https://exact.cs.fau.de/api/v1/ and docu
- REST-API Client https://github.com/ChristianMarzahl/EXACT-Sync
- Dynamic OpenAPI client generation with Swagger and EXACT-API.yml
- Sync with the offline Tool SlideRunner
- team creation
- upload image sets in the multiple formats like: whole slide image (WSI) formats or .png, .jpg, .jepg, .bmp etc.
- bounding box, circle and polygon support
- export format creation
- label export
- image preloading for labeling and verification
- label verification
- upload of existing labels
- WSI viewer
- Caching
- Registration Registration.md
Documentation
Issues with the notebooks on GitHub? Please use NBViewer
| Describtion | Video |
|---|---|
| EXACT installation with Docker (en) |  |
| EXACT First steps (en) |  |
| EXACT Product and Annotationtype defintion (en) |  |
| Study and annotation modes (en) |  <br> Code <br> Code |
| AnnotationMaps (en) |  <br> Code <br> Code |
| DensityMaps (en) |  <br> Code <br> Code |
| Cluster annotations (en) |  <br> Code <br> Code |
| Cluster sounds (en) |  |
| AnnotationVersioning (en) | 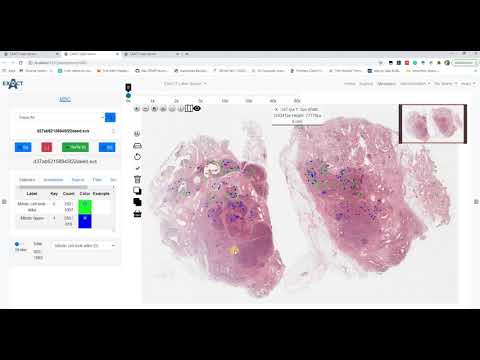 |
| Inference (en) | 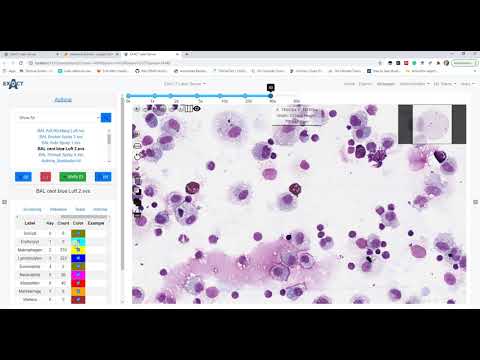 <br> [Code](doc/Inference Asthma.ipynb) <br> [Code](doc/Inference Asthma.ipynb) |
| Segmentation (en) | 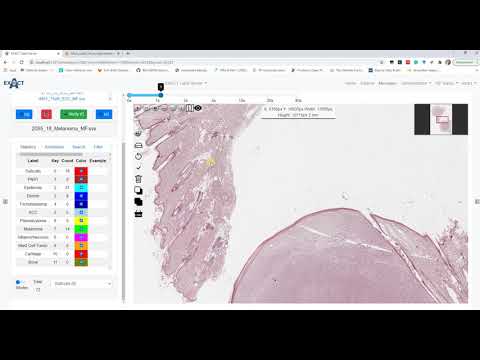 <br> Code <br> Code |
| EXACT Media Files (en) | 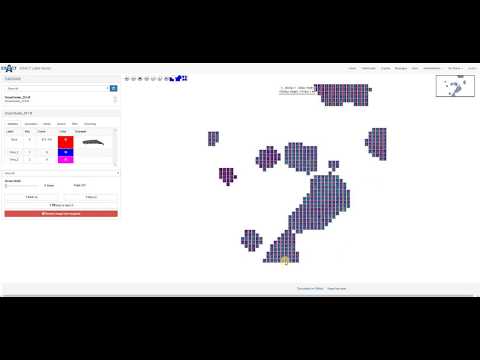 |
| Datasets (en) | 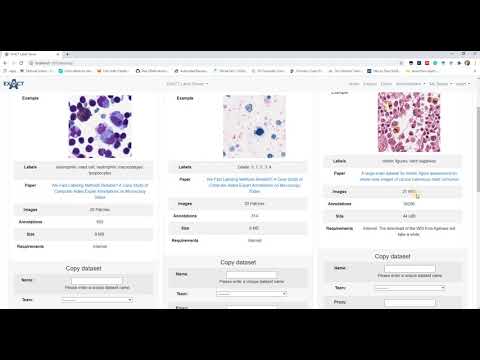 |
| Explains how teams, products and annotation types are created (de) | 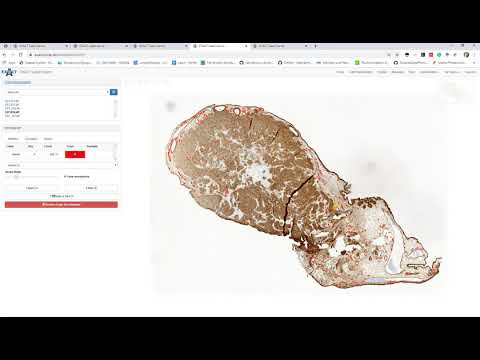 |
| Describes how to create, view and edit image sets and upload images. (en) | 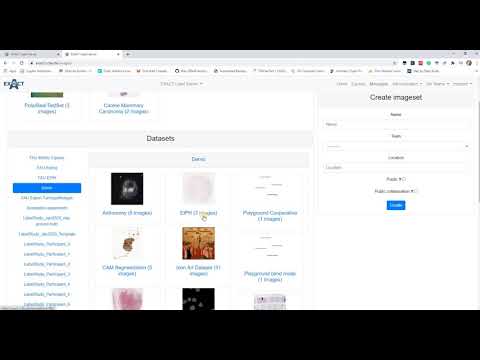 |
| Describes basic viewer and plugin functions. (en) | 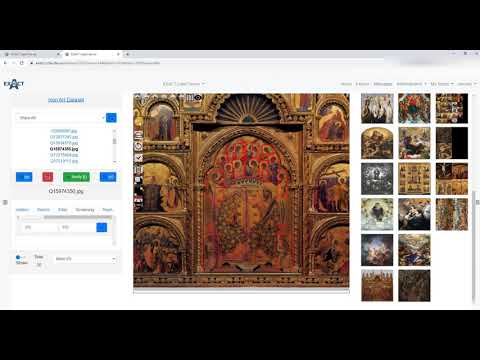 |
| Describes the image registration functionality | 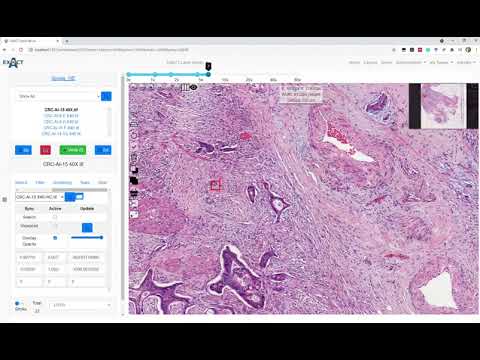 |
| Explains collaboratory annotation features (de) | 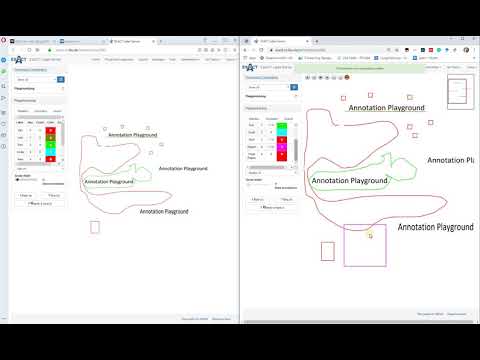 |
| Shows multiple types of datasets (de) |  |
| Explains the screening plugin for WSI (de) | 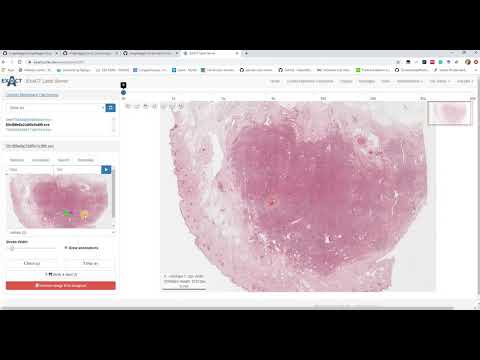 |
| Syncronisation with the offline tool SlideRunner (en) | 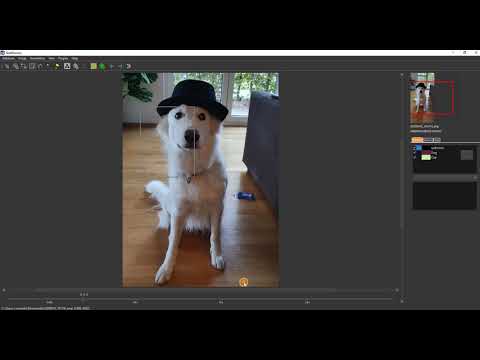 |
| Advanced polygon annotation operations (de) |  |
| REST-API Example | pip install EXCAT-Sync <br> Code <br> Notebooks <br> Browsable-API |
Install
Docker
Install Docker
Checkout the latest release:
git clone https://github.com/DeepMicroscopy/Exact.git
Production
Additional features:
- gunicorn
- nginx
Copy the files env.dev and env.dev.db, rename to env.prod and env.prod.db and change settings according to your preferences.
Copy and rename settings.py.example to settings.py in the exact folder.
Enable caching in the settings.py according to the documentation.
Build and run the container:
docker-compose -f docker-compose.prod.yml up -d --build
docker-compose -f docker-compose.prod.yml exec web python3 manage.py migrate --noinput
docker-compose -f docker-compose.prod.yml exec web python3 manage.py createsuperuser
docker-compose -f docker-compose.prod.yml exec web python3 manage.py collectstatic --no-input --clear
docker-compose -f docker-compose.prod.yml logs -f
Navigate to http://localhost:1337/
Development
Copy and rename settings.py.example to settings.py in the exact folder.
Modify the configuration files: env.dev and env.dev.db or use the default configuration.
Build and run the container:
docker-compose -f docker-compose.yml up -d --build
docker-compose logs -f
Navigate to http://localhost:8000/ For default the super user login is:
User: exact
Pw: exact
Cloud
To use cloud services like amazon fargate. The exact server has to connect to a cloud database like amazon rds.
Copy the files env.dev and env.dev.db, rename to env.prod and env.prod.aws-db and change settings according to your preferences.
Copy and rename settings.py.example to settings.py in the exact folder.
Build and run the container:
docker-compose -f docker-compose.prod.aws-db.yml up -d --build
docker-compose -f docker-compose.prod.aws-db.yml exec web python3 manage.py migrate --noinput
docker-compose -f docker-compose.prod.aws-db.yml exec web python3 manage.py createsuperuser
docker-compose -f docker-compose.prod.aws-db.yml exec web python3 manage.py collectstatic --no-input --clear
docker-compose -f docker-compose.prod.aws-db.yml logs -f
Send container to AWS ECR
Invoke-Expression -Command (aws ecr get-login --no-include-email)
docker tag exact_nginx:latest **************.dkr.ecr.eu-central-1.amazonaws.com/exact_nginx:latest
docker tag exact_web:latest **************.dkr.ecr.eu-central-1.amazonaws.com/exact:latest
docker push **************.dkr.ecr.eu-central-1.amazonaws.com/exact_nginx:latest
docker push **************.dkr.ecr.eu-central-1.amazonaws.com/exact:latest
Windows
The Server is also runnig on Windows but I would recommend to use docker in that case.
MacOS or Linux
Ubuntu20.04 has a known issue with openslide. Fix: Build pixman
Install libvips and verify by executing the comand vips:
https://libvips.github.io/libvips/
[Ubuntu](https://github.com/libvips/libvips/wiki/Build-for-Ubuntu)
Checkout the latest release:
git clone https://github.com/DeepMicroscopy/Exact.git
In our production Senty is used for error reporting (pip install raven). django-auth-ldap is used for login via ldap uwsgi is used to serve the app to nginx
Install Python Dependencies:
pip3 install -r requirements.txt
Copy settings.py.example to settings.py in the exact folder:
cp exact/exact/settings.py.example exact/exact/settings.py
and customize the settings.py.
The following settings should probably be changed:
- The secret key
- The DEBUG setting
- The ALLOWED_HOSTS
- The database settings
- The UPLOAD_FS_GROUP to the id of the group that should access and create the uploaded images
For the database, postgresql is used. Install it by running sudo apt install postgresql
You will also need opencv3 and other packages. You can install that by running apt-get update && apt-get install python3-pip dos2unix python3-openslide python3-opencv libvips libvips-dev
Initialize the database cluster with sudo -iu postgres initdb --locale en_US.UTF-8 -D '/var/lib/postgres/data'.
Note: It may be that initdb is not in your current PATH (seems to be default for postgresql >= 10), in this case, you have to specify the proper path to initdb, e.g:
sudo -iu postgres /usr/lib/postgresql/*/bin/initdb --locale en_US.UTF-8 -D '/var/lib/postgresql/data' (for Ubuntu systems)
To start the postgresql server, run sudo systemctl start postgresql.service. If the server should always be started on boot, run sudo systemctl enable postgresql.service.
Then, create the user and the database by running
sudo -iu postgres psql
and then, in the postgres environment
CREATE USER exact PASSWORD 'exact';
CREATE DATABASE exact WITH OWNER exact ENCODING UTF8;
where of course the password and the user should be adapted to the ones specified in the database settings in the settings.py.
To initialize the database, run python3 manage.py migrate
To create an administrator user, run python3 manage.py createsuperuser.
python3 manage.py runserver starts the server with the configuration given in the settings.py file.
To create annotation types, log into the application and click on Administration at the very bottom of the home page.
For production systems it is necessary to run the following commands after each upgrade
python3 manage.py migrate
python3 manage.py compilemessages
python3 manage.py collectstatic
Our production uwisgi config is
[uwsgi]
socket = /tmp/exact.socket
chmod-socket = 666
chdir = /srv/exact/Exact/exact/
master = true
binary-path = /usr/bin/uwsgi
virtualenv = /srv/exact/virtualenv/exact
module = exact.wsgi
uid = exact
gid = exact
processes = 6
#async = 10
threads = 1
#logto = /var/log/exact.log
plugins = python3,logfile
logger = file:/var/log/exact.log
Example Nginx Config:
upstream exact {
server web:8000;
}
server {
listen 80;
client_max_body_size 10000M;
keepalive_timeout 65;
proxy_connect_timeout 6000s;
proxy_send_timeout 6000s;
proxy_read_timeout 6000s;
send_timeout 6000s;
client_body_timeout 6000s;
location / {
proxy_pass http://exact;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_set_header Host $host;
proxy_redirect off;
}
location /static/ {
expires 1h;
alias /home/app/web/static/;
}
location /media/ {
expires 1h;
alias /home/app/web/media/;
}
}
Verification E-Mails
Please add credentials to the settings.py
EMAIL_BACKEND = 'django.core.mail.backends.smtp.EmailBackend'
EMAIL_HOST = 'smtp.googlemail.com' # for example gmail
EMAIL_PORT = '587'
EMAIL_HOST_USER = 'example@gmail.com'
EMAIL_HOST_PASSWORD = 'example'
EMAIL_USE_TLS = True
EMAIL_USE_SSL = False
Upgrade
pip install -U -r requirements.txt
python3 manage.py migrate
for additional steps on some releases see instructions in UPGRADE.md
Key shortcuts
Viewer
| Key | Modifier | Function |
|---|---|---|
| Del,x | Delete annotation | |
| Escape | Cancel editing | |
| Enter | Confirm / Save | |
| Ctrl + z | Undo | |
| c | Toggle annotation mode | |
| y | Toggle annotation visibility | |
| b | Push currently selected annotation type into the background | |
| Ctrl,a | Draw annotation on top of existing one | |
| 0,1,2,3,4 | Change label of local annotations | |
| 0,1,2,3,4 | Shift | Change label of global annotations |
| q | Previous image | |
| q | Shift | Previous frame |
| e | Next image | |
| e | Shift | Next frame |
| r | Rotate image (Warning: annotations are not affected) | |
| f | Flip image (Warning: annotations are not affected) | |
| s | Scissor (Delete from selected object) | |
| g | Glue (Add to selected object) | |
| d | Knife (Draw a line to split objects) | |
| Mouse wheel | Shift | Paint brush |
| Arrow keys | Move viewing window |
Screening Viewer
| Key | Modifier | Function |
|---|---|---|
| a | screen left tile | |
| w | screen up tile | |
| s | screen down tile | |
| d | screen right tile | |
| j | navigate to left tile | |
| i | navigate to up tile | |
| k | navigate to down tile | |
| l | navigate to right tile |
Sync Plugin
| Key | Modifier | Function |
|---|---|---|
| o | toggle visibility |
REST-API
OpenAPI-Schema
GET /api/v1/openapi
Examples
https://github.com/rsinger86/drf-flex-fields
https://django-filter.readthedocs.io/en/master/
Token
$ curl -X POST -d '{"username": "exact","password": "top_secret"}' -H
'Content-Type: application/json' http://127.0.0.1:8000/api/auth/token/login/
All suported classes
GET /api/v1/
{
"users/users": "http://127.0.0.1:8000/api/v1/users/users/",
"users/teams": "http://127.0.0.1:8000/api/v1/users/teams/",
"users/team_membership": "http://127.0.0.1:8000/api/v1/users/team_membership/",
"images/images": "http://127.0.0.1:8000/api/v1/images/images/",
"images/image_sets": "http://127.0.0.1:8000/api/v1/images/image_sets/",
"images/set_tags": "http://127.0.0.1:8000/api/v1/images/set_tags/",
"images/screening_modes": "http://127.0.0.1:8000/api/v1/images/screening_modes/",
"annotations/annotations": "http://127.0.0.1:8000/api/v1/annotations/annotations/",
"annotations/annotation_types": "http://127.0.0.1:8000/api/v1/annotations/annotation_types/",
"annotations/annotation_media_files": "http://127.0.0.1:8000/api/v1/annotations/annotation_media_files/",
"annotations/verifications": "http://127.0.0.1:8000/api/v1/annotations/verifications/",
"annotations/log_image_actions": "http://127.0.0.1:8000/api/v1/annotations/log_image_actions/",
"administration/products": "http://127.0.0.1:8000/api/v1/administration/products/"
}
Filter all image_sets with "Katze" in name and expand products and main_annotation_type
GET /api/v1/images/image_sets/?name__contains=Katze&expand=product_set,main_annotation_type
{
"count": 1,
"next": null,
"previous": null,
"results": [
{
"id": 3,
"name": "EIPH-Katze",
"path": "exact_1_3",
"location": null,
"description": "",
"images": [
17,
20,
22,
23,
26,
27,
30
],
"product_set": [
{
"id": 2,
"name": "EIPH",
"description": "",
"team": 1,
"creator": 1,
"imagesets": [
2,
3,
16
],
"annotationtype_set": [
10,
11,
12,
13,
14
]
}
],
"main_annotation_type": {
"id": 10,
"name": "0",
"vector_type": 1,
"node_count": 0,
"enable_concealed": false,
"enable_blurred": false,
"color_code": "#0000FF",
"default_width": 120,
"default_height": 120,
"sort_order": 0,
"closed": true,
"area_hit_test": true,
"product": 2
},
"set_tags": [],
"team": 1,
"creator": 1
}
]
}
Include(fields) or Exclude(omit) fields
GET /api/v1/images/image_sets/?name__contains=Katze&fields=name
{
"count": 1,
"next": null,
"previous": null,
"results": [
{
"name": "EIPH-Katze"
}
]
}
GET /api/v1/images/image_sets/?name__contains=Katze&omit=images,product_set
{
"count": 1,
"next": null,
"previous": null,
"results": [
{
"id": 3,
"name": "EIPH-Katze",
"path": "exact_1_3",
"location": null,
"description": "",
"main_annotation_type": 10,
"set_tags": [],
"team": 1,
"creator": 1
}
]
}
Used dependencies
The exact relies on the following plugins, libraries and frameworks:
| Name | Version | License |
|---|---|---|
| Django | 3.0 | BSD |
| Pillow | 5.4.1 | Standard PIL License |
| asgiref | 3.2.3 | BSD |
| confusable-homoglyphs | 3.2.0 | MIT |
| django-friendly-tag-loader | 1.3.1 | MIT |
| django-registration | 3.0.1 | MIT |
| django-widget-tweaks | 1.4.3 | MIT license |
| djangorestframework | 3.11.0 | BSD |
| fasteners | 0.14.1 | ASL 2.0 |
| gunicorn | 19.9.0 | MIT |
| imagecodecs-lite | 2019.12.3 | BSD |
| monotonic | 1.5 | Apache |
| numpy | 1.17.4 | BSD |
| opencv-python | 4.1.2.30 | MIT |
| openslide-python | 1.1.1 | GNU Lesser General Public License, version 2.1 |
| psycopg2-binary | 2.7.7 | LGPL with exceptions or ZPL |
| pytz | 2018.9 | MIT |
| six | 1.12.0 | MIT |
| sqlparse | 0.3.0 | BSD |
| tifffile | 2019.7.26.2 | BSD |
| Bootstrap | 4.4 | BSD |
| jQuery | 3.4.1 | MIT |
| jQuery-Autocomplete | 1.4.1 | |
| jQuery-File-Upload | 10.7.0 | MIT |
| OpenSeadragon | 2.4.1 | BSD-3 |
We are grateful to the maintainers and contributors of the respective projects.
ImageTagger Reference
This paper describes the Bit-Bots imagetagger we build on in depth. Please cite if you use this tool in your research:
FIEDLER, Niklas, et al. imagetagger: An Open Source Online Platform for Collaborative Image Labeling. In: RoboCup 2018: Robot World Cup XXII. Springer, 2018.
@inproceedings{imagetagger2018,
author={Fiedler, Niklas and Bestmann, Marc and Hendrich, Norman},
year={2018},
title={Imagetagger: An Open Source Online Platform for Collaborative Image Labeling},
booktitle={RoboCup 2018: Robot World Cup XXII},
organization={Springer}
}
