Awesome
<div align="center"> <br /> <p> <a href="https://github.com/Chokyotager/NotYetAnotherNightshade"><img src="/art/NYAN.png" alt="banner" /></a> </p> <br /> <p> </p> </div>About
NotYetAnotherNightshade (NYAN) is a graph variational encoder as described in the manuscript "Application of variational graph encoders as an effective generalist algorithm in holistic computer-aided drug design".
It allows for the embedding of molecules into a continuous latent space, and subsequent surrogate model training for molecular property prediction not limited to drug design and other chemistry applications.
The latent space method as described can also be used to perform highly accelerated, very high throughput virtual screening for computer-aided drug discovery of up to a few billion compounds.
This repository contains the code we used in training of new encoders, construction of surrogate models, and latent space potentiation as described in the manuscript. We have also included utility tools for decoding and encoding molecules, so that you can fit and train your own surrogate models.
Paper
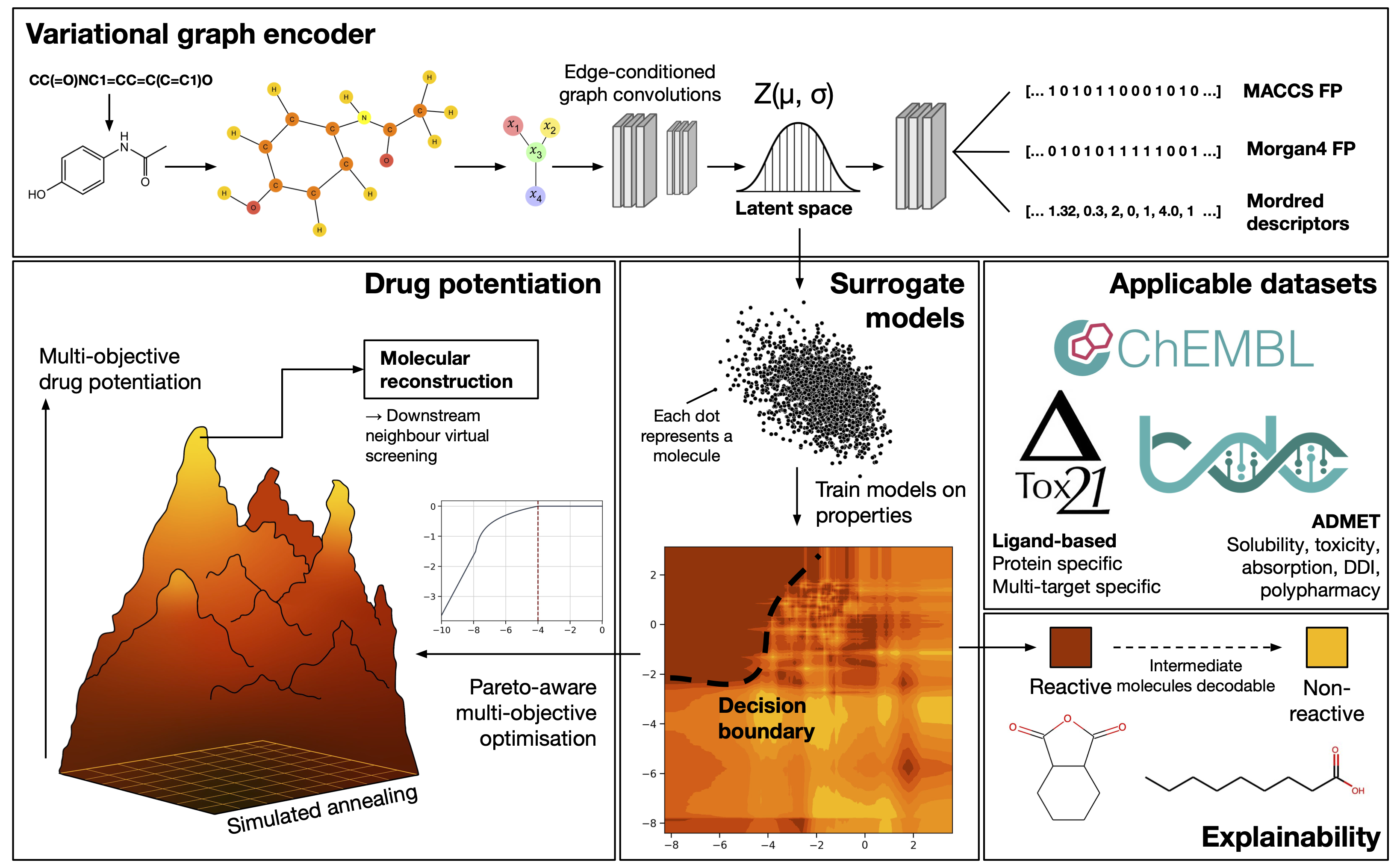 Please read the paper for more details:
Please read the paper for more details:
Publication: https://www.nature.com/articles/s42256-023-00683-9
Publication DOI: https://doi.org/10.1038/s42256-023-00683-9
Preprint: https://www.biorxiv.org/content/10.1101/2023.01.11.523575v1
Preprint DOI: https://doi.org/10.1101/2023.01.11.523575
Installation
git clone https://github.com/Chokyotager/NotYetAnotherNightshade.git
cd NotYetAnotherNightshade
conda env create -f environment.yml
conda activate nyan
There is also an explicit link file in requirements.txt for all Conda packages.
NYAN has been tested on two separate Ubuntu 22.04.1 LTS (GNU/Linux 5.15.0-58-generic x86_64) systems. It theoretically should work for any environment so long as all package requirements are fulfilled.
Installation should take under ten minutes in most cases.
Practical demonstration!
There is an IPython Notebook that you can open using Jupyter and/or other notebooks (not tested) named NYAN_demo.ipynb. It contains steps on how to download the data, train it, and then potentiate molecules.
Thank you very much to the (currently) anonymous reviewer for suggesting this!
Usage
By default, the saved model ZINC-extmodel5hk-3M is used. This is the same model which was used throughout the manuscript to generate the figures. You should be able to reproduce the results accordingly.
If you want to train your own model, look at the "training your own model" section below.
Conversion of SMILES to latent space
This tool converts any SMILES molecule into a continuous mathematical space of 64 dimensions.
Please use "encode_smiles.py".
python3 encode_smiles.py <input SMILES file> <output file>
The output will be tab-delimited. You can then use this latent space to train surrogate models to your liking. I.e. you can use SKLearn to build ExtraTrees regressors/classifiers to predict ADMET properties.
You can look into datasets/example.smi for an example input file.
The runtime per molecule should not exceed 10 seconds after program initialisation. However, faster GPUs (such as the Nvidia RTX 3090®) that we tested this on can process up to 500 molecules per second.
Conversion of latent space into molecular fingerprints
This tool does the opposite and converts a 64-dimensional latent space into molecular fingerprints and Mordred descriptors.
python3 decode_latent.py <input latent TSV> <output file>
The first column of the input latent TSV is treated as an ID. Subsequent columns are of the latent space. In total, there should be 65 columns in your input file. All elements should be delimited by tabs. An example of an input file that you can feed in here is the output from encode_smiles.py.
The output will be tab-delimited. You can use this to infer molecular properties or match against a known fingerprint database.
You can look into datasets/example_latents.tsv for an example input file.
The runtime per molecule should not exceed 10 seconds after program initialisation.
Training your own encoder
If you want to train your own model, please edit config.json.
You can then train your model using python3 train.py
Other experiments done in the paper
We haven't gotten around to build user-friendly tools for molecular potentiation yet. The code that was used in the paper that was used to do this can be found in misc-code/NYAN-potentiator.
We have also added one in misc-code/referfence-encoding-decoding for latent space and FP searching. To use the stuff in misc-code/, you would probably have to change the code directly.
Contribution
Users are welcome to contribute to the development of the model through pull-requests.
Our future direction is to make this tool as user-friendly as possible, and to release individual surrogate models that we have trained. Please continue to check back on this repo for the latest updates!
Maintenance
The current project is maintained by Hilbert Lam and Robbe Pincket. Correspondence can be found in the manuscript. You can also contact Hilbert / Robbe via email there or (informally) on Discord, using the handles ChocoParrot#8925, Kroppeb#2845 respectively (we're chill people).
Reproduction of results in the paper
Because we used a large variety of datasets (each with different licenses), we are unable to clone these datasets into this repository. You can reproduce the results we obtained by downloading the respective datasets, and converting them into latent vectors using NYAN.
The latent spaces can then be trained and evaluated accordingly using methods from SKLearn or any other applicable machine learning library.
License
License details can be found in the LICENSE file.
Citation
@article{lam_application_2023,
title = {Application of variational graph encoders as an effective generalist algorithm in computer-aided drug design},
issn = {2522-5839},
url = {https://www.nature.com/articles/s42256-023-00683-9},
doi = {10.1038/s42256-023-00683-9},
language = {en},
urldate = {2023-07-11},
journal = {Nature Machine Intelligence},
author = {Lam, Hilbert Yuen In and Pincket, Robbe and Han, Hao and Ong, Xing Er and Wang, Zechen and Hinks, Jamie and Wei, Yanjie and Li, Weifeng and Zheng, Liangzhen and Mu, Yuguang},
month = jul,
year = {2023},
}